2024

Hotspot-Driven Peptide Design via Multi-Fragment Autoregressive Extension
Jiahan Li*, Tong Chen*, Shitong Luo, Chaoran Cheng, Jiaqi Guan, Ruihan Guo, Sheng Wang, Ge Liu, Jian Peng, Jianzhu Ma (* equal contribution)
Submitted to ICLR 2025
PepHAR introduces a hot-spot-driven autoregressive generative model for peptide design, combining energy-based hot spot sampling, fragment-based extension, and iterative refinement to generate structurally valid peptides tailored to specific protein targets, demonstrating strong potential in computational peptide binder design.
Hotspot-Driven Peptide Design via Multi-Fragment Autoregressive Extension
Jiahan Li*, Tong Chen*, Shitong Luo, Chaoran Cheng, Jiaqi Guan, Ruihan Guo, Sheng Wang, Ge Liu, Jian Peng, Jianzhu Ma (* equal contribution)
Submitted to ICLR 2025
PepHAR introduces a hot-spot-driven autoregressive generative model for peptide design, combining energy-based hot spot sampling, fragment-based extension, and iterative refinement to generate structurally valid peptides tailored to specific protein targets, demonstrating strong potential in computational peptide binder design.
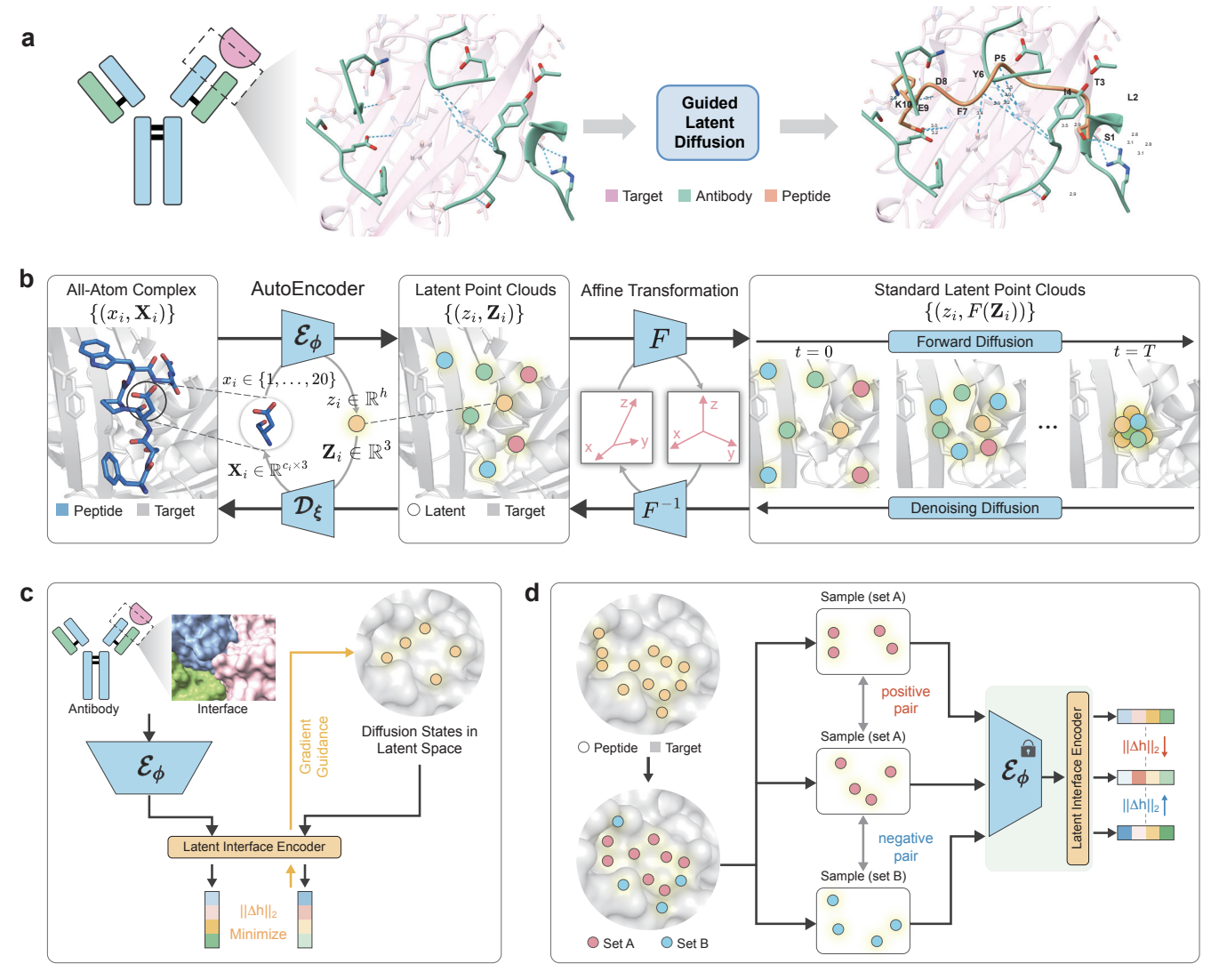
All-Atom Peptide Design by Mimicking Binding Interface
Xiangzhe Kong, Rui Jiao, Haowei Lin, Ruihan Guo, Wenbing Huang, Wei-Ying Ma, Zihua Wang, Yang Liu, Jianzhu Ma
Submitted to Nature Methods, under review. 2024
PepMimic is an machine learning model for designing peptide drug candidates by mimicking binding interfaces, achieving dissociation constants as low as $10^{-9}$M with a success rate 20,000 times higher than random screening, and demonstrating therapeutic potential through extensive cellular and in vivo validations.
All-Atom Peptide Design by Mimicking Binding Interface
Xiangzhe Kong, Rui Jiao, Haowei Lin, Ruihan Guo, Wenbing Huang, Wei-Ying Ma, Zihua Wang, Yang Liu, Jianzhu Ma
Submitted to Nature Methods, under review. 2024
PepMimic is an machine learning model for designing peptide drug candidates by mimicking binding interfaces, achieving dissociation constants as low as $10^{-9}$M with a success rate 20,000 times higher than random screening, and demonstrating therapeutic potential through extensive cellular and in vivo validations.
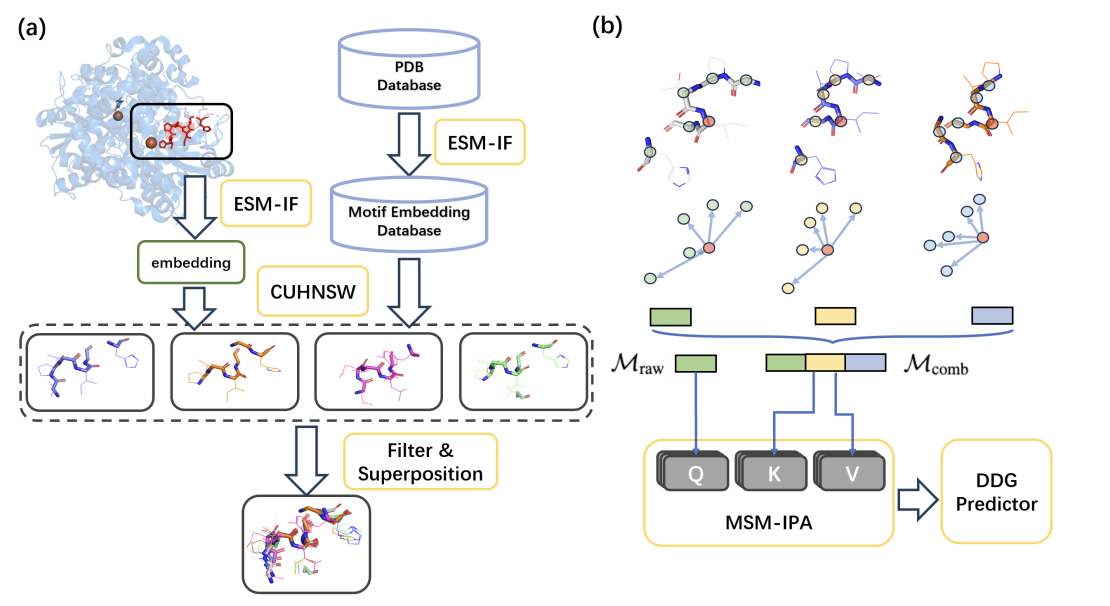
Enhancing Protein Mutation Effect Prediction through a Retrieval-Augmented Framework
Ruihan Guo*, Rui Wang*, Ruidong Wu*, Zhizhou Ren, Jiahan Li, Shitong Luo, Zuofan Wu, Qiang Liu, Jian Peng, Jianzhu Ma (* equal contribution)
NeurIPS 2024
This work introduces a retrieval-augmented framework that incorporates similar local structural motifs from a pre-trained protein structure encoder, achieving state-of-the-art performance in protein mutation effect prediction and providing a scalable solution for studying mutation impacts.
Enhancing Protein Mutation Effect Prediction through a Retrieval-Augmented Framework
Ruihan Guo*, Rui Wang*, Ruidong Wu*, Zhizhou Ren, Jiahan Li, Shitong Luo, Zuofan Wu, Qiang Liu, Jian Peng, Jianzhu Ma (* equal contribution)
NeurIPS 2024
This work introduces a retrieval-augmented framework that incorporates similar local structural motifs from a pre-trained protein structure encoder, achieving state-of-the-art performance in protein mutation effect prediction and providing a scalable solution for studying mutation impacts.
ME-PATS: Mutually Enhancing Search-Based Planner and Learning-Based Agent for TractorTrailer Systems
Ke Fan, Zhizhou Ren, Ruihan Guo, Jinpeng Zhang, Zhuo Huang, Yuan Zhou, Zufeng Zhang
Submitted to ICRA 2025
ME-PATS introduces a simulator for tractor-trailer systems and integrates search-based A* planning with reinforcement learning to address real-world maneuvering challenges.
ME-PATS: Mutually Enhancing Search-Based Planner and Learning-Based Agent for TractorTrailer Systems
Ke Fan, Zhizhou Ren, Ruihan Guo, Jinpeng Zhang, Zhuo Huang, Yuan Zhou, Zufeng Zhang
Submitted to ICRA 2025
ME-PATS introduces a simulator for tractor-trailer systems and integrates search-based A* planning with reinforcement learning to address real-world maneuvering challenges.
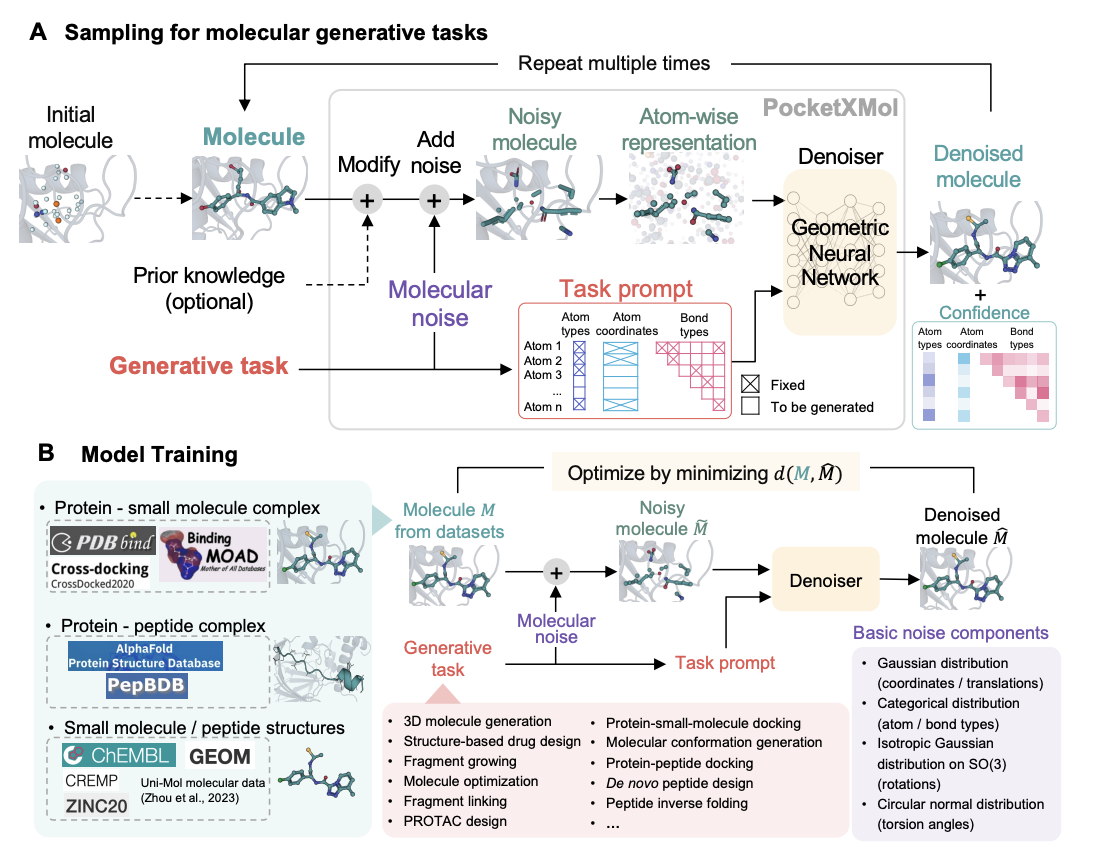
Decipher Fundamental Atomic Interactions to Unify Generative Molecular Docking and Design
Xingang Peng, Ruihan Guo, Yan Xu, Jiaqi Guan, Yinjun Jia, Yanwen Huang, Muhan Zhang, Jian Peng, Jiayu Sun, Chuanhui Han, Zihua Wang, Jianzhu Ma
Submitted to Cell, under review. 2024
PocketXMol is an all-atom model that unifies diverse molecular tasks under a single framework without fine-tuning, excelling in small molecule and peptide design, with demonstrated success in creating caspase-9 inhibitors and PD-L1-binding peptides validated in cellular and animal models.
Decipher Fundamental Atomic Interactions to Unify Generative Molecular Docking and Design
Xingang Peng, Ruihan Guo, Yan Xu, Jiaqi Guan, Yinjun Jia, Yanwen Huang, Muhan Zhang, Jian Peng, Jiayu Sun, Chuanhui Han, Zihua Wang, Jianzhu Ma
Submitted to Cell, under review. 2024
PocketXMol is an all-atom model that unifies diverse molecular tasks under a single framework without fine-tuning, excelling in small molecule and peptide design, with demonstrated success in creating caspase-9 inhibitors and PD-L1-binding peptides validated in cellular and animal models.
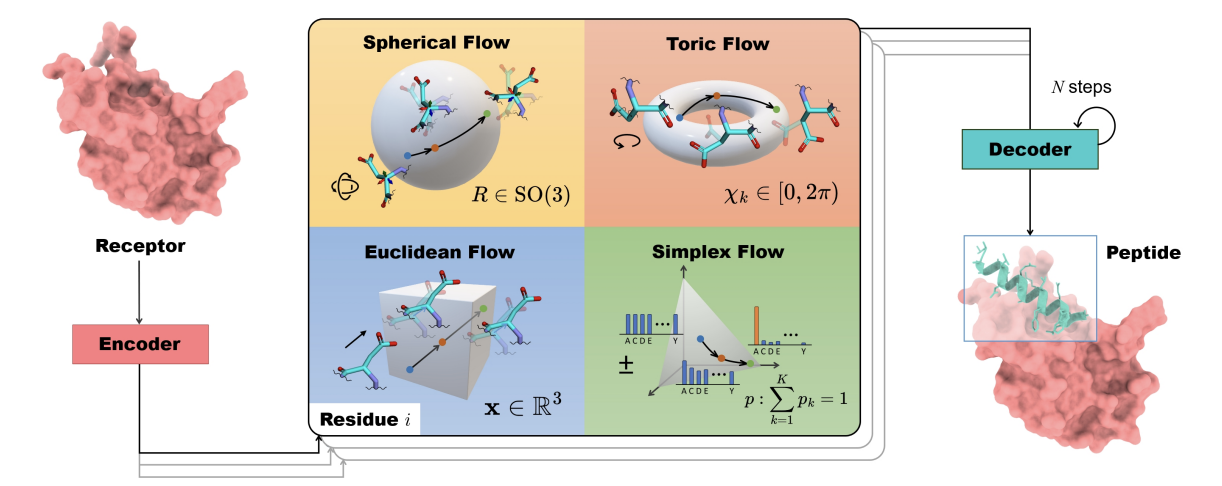
Full-Atom Peptide Design based on Multi-modal Flow Matching
Jiahan Li*, Chaoran Cheng*, Zuofan Wu, Ruihan Guo, Shitong Luo, Zhizhou Ren, Jian Peng, Jianzhu Ma (* equal contribution)
ICML 2024
PepFlow introduces a multimodal deep generative model based on the flow-matching framework for full-atom peptide design, leveraging SE(3) manifolds and high-dimensional tori to model backbone orientations and side-chain dynamics, achieving state-of-the-art performance across peptide design tasks.
Full-Atom Peptide Design based on Multi-modal Flow Matching
Jiahan Li*, Chaoran Cheng*, Zuofan Wu, Ruihan Guo, Shitong Luo, Zhizhou Ren, Jian Peng, Jianzhu Ma (* equal contribution)
ICML 2024
PepFlow introduces a multimodal deep generative model based on the flow-matching framework for full-atom peptide design, leveraging SE(3) manifolds and high-dimensional tori to model backbone orientations and side-chain dynamics, achieving state-of-the-art performance across peptide design tasks.
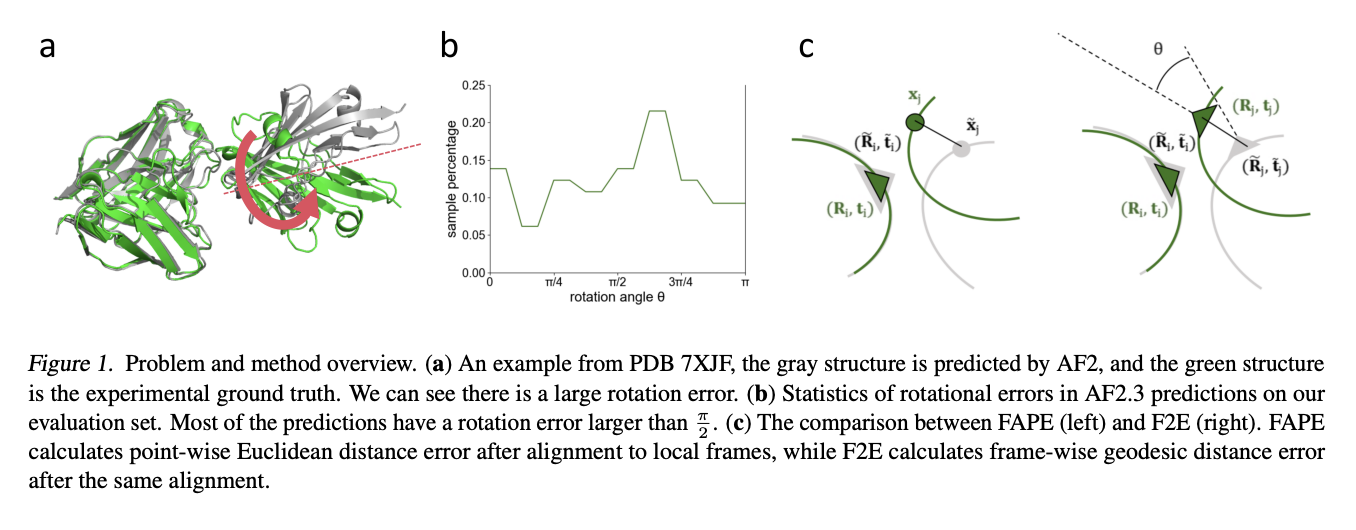
FAFE: Immune Complex Modeling with Geodesic Distance Loss on Noisy Group Frames
Ruidong Wu*, Ruihan Guo*, Rui Wang*, Shitong Luo, Yue Xu, Jiahan Li, Jianzhu Ma, Qiang Liu, Yunan Luo, Jian Peng (* equal contribution)
ICML 2024 Spotlight
This work introduces Frame Aligned Frame Error (FAFE), a novel geodesic loss that overcomes AlphaFold2's gradient vanishing issue in high-rotational-error targets, enabling more accurate antibody-antigen complex modeling and achieving up to a 182% improvement in correct docking rates.
FAFE: Immune Complex Modeling with Geodesic Distance Loss on Noisy Group Frames
Ruidong Wu*, Ruihan Guo*, Rui Wang*, Shitong Luo, Yue Xu, Jiahan Li, Jianzhu Ma, Qiang Liu, Yunan Luo, Jian Peng (* equal contribution)
ICML 2024 Spotlight
This work introduces Frame Aligned Frame Error (FAFE), a novel geodesic loss that overcomes AlphaFold2's gradient vanishing issue in high-rotational-error targets, enabling more accurate antibody-antigen complex modeling and achieving up to a 182% improvement in correct docking rates.
2022
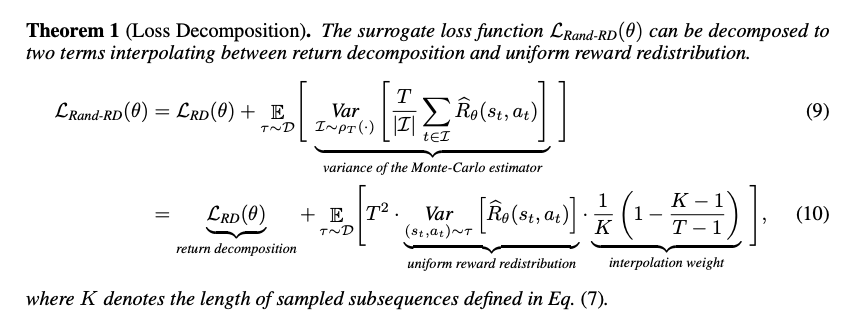
Learning Long-Term Reward Redistribution via Randomized Return Decomposition
Zhizhou Ren, Ruihan Guo, Yuan Zhou, Jian Peng
ICLR 2022 Spotlight
This work addresses episodic reinforcement learning with trajectory feedback by introducing Randomized Return Decomposition (RRD), a reward redistribution algorithm that uses Monte-Carlo sampling to scale least-squares-based proxy reward learning for long-horizon tasks, achieving significant improvements over baseline methods.
Learning Long-Term Reward Redistribution via Randomized Return Decomposition
Zhizhou Ren, Ruihan Guo, Yuan Zhou, Jian Peng
ICLR 2022 Spotlight
This work addresses episodic reinforcement learning with trajectory feedback by introducing Randomized Return Decomposition (RRD), a reward redistribution algorithm that uses Monte-Carlo sampling to scale least-squares-based proxy reward learning for long-horizon tasks, achieving significant improvements over baseline methods.